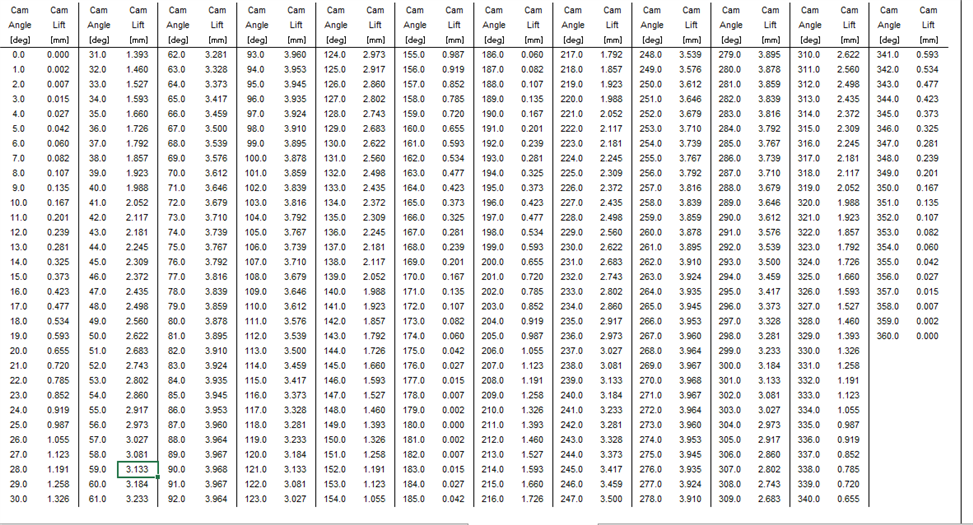
 We're wondering how to import this data into the demon.
We're wondering how to import this data into the demon.
Your Products have been synced, click here to refresh
Your Products have been synced, click here to refresh
 We're wondering how to import this data into the demon.
We're wondering how to import this data into the demon.
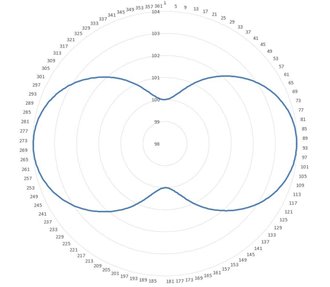
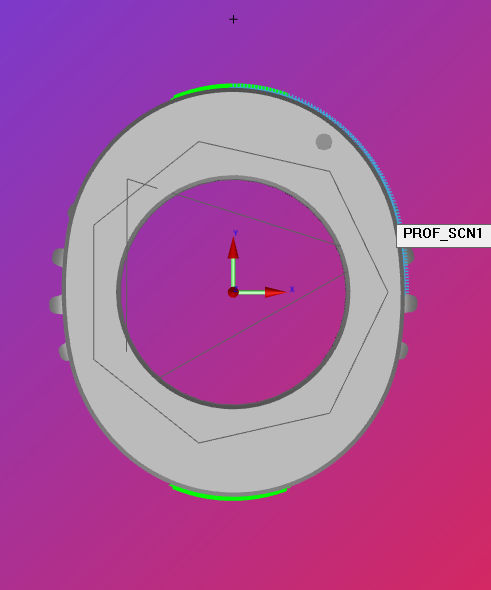
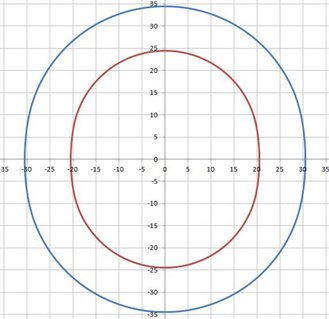
OF_SCN1 =FEAT/SCAN,FREEFORM,NUMBER OF HITS=91,SHOW HITS=NO,SHOWALLPARAMS=NO
MEAS/SCAN
BASICSCAN/FREEFORM,NUMBER OF HITS=91,SHOW HITS=YES,SHOWALLPARAMS=NO
HIT/VECTOR,<20.5,0,0>,<1,0,0>,<20.5,0,0>,T=0
HIT/VECTOR,<20.4986,0.3578,0>,<0.9998477,0.0174524,0>,<20.4986,0.3578,0>,T=0
HIT/VECTOR,<20.4942,0.7157,0>,<0.9993908,0.0348995,0>,<20.4942,0.7157,0>,T=0
HIT/VECTOR,<20.4869,1.0737,0>,<0.9986295,0.052336,0>,<20.4869,1.0737,0>,T=0
HIT/VECTOR,<20.4767,1.4319,0>,<0.9975641,0.0697565,0>,<20.4767,1.4319,0>,T=0
HIT/VECTOR,<20.4636,1.7903,0>,<0.9961947,0.0871557,0>,<20.4636,1.7903,0>,T=0
HIT/VECTOR,<20.4475,2.1491,0>,<0.9945219,0.1045285,0>,<20.4475,2.1491,0>,T=0
HIT/VECTOR,<20.4283,2.5083,0>,<0.9925462,0.1218693,0>,<20.4283,2.5083,0>,T=0
HIT/VECTOR,<20.4062,2.8679,0>,<0.9902681,0.1391731,0>,<20.4062,2.8679,0>,T=0
HIT/VECTOR,<20.3809,3.228,0>,<0.9876883,0.1564345,0>,<20.3809,3.228,0>,T=0
HIT/VECTOR,<20.3526,3.5887,0>,<0.9848078,0.1736482,0>,<20.3526,3.5887,0>,T=0
HIT/VECTOR,<20.321,3.95,0>,<0.9816272,0.190809,0>,<20.321,3.95,0>,T=0
HIT/VECTOR,<20.2862,4.312,0>,<0.9781476,0.2079117,0>,<20.2862,4.312,0>,T=0
HIT/VECTOR,<20.2481,4.6747,0>,<0.9743701,0.2249511,0>,<20.2481,4.6747,0>,T=0
HIT/VECTOR,<20.2066,5.0381,0>,<0.9702957,0.2419219,0>,<20.2066,5.0381,0>,T=0
HIT/VECTOR,<20.1616,5.4023,0>,<0.9659258,0.258819,0>,<20.1616,5.4023,0>,T=0
HIT/VECTOR,<20.1129,5.7673,0>,<0.9612617,0.2756374,0>,<20.1129,5.7673,0>,T=0
HIT/VECTOR,<20.0606,6.1331,0>,<0.9563048,0.2923717,0>,<20.0606,6.1331,0>,T=0
HIT/VECTOR,<20.0044,6.4998,0>,<0.9510565,0.309017,0>,<20.0044,6.4998,0>,T=0
HIT/VECTOR,<19.9442,6.8673,0>,<0.9455186,0.3255682,0>,<19.9442,6.8673,0>,T=0
HIT/VECTOR,<19.8797,7.2356,0>,<0.9396926,0.3420201,0>,<19.8797,7.2356,0>,T=0
HIT/VECTOR,<19.8102,7.6044,0>,<0.9335804,0.358368,0>,<19.8102,7.6044,0>,T=0
HIT/VECTOR,<19.7352,7.9736,0>,<0.9271839,0.3746066,0>,<19.7352,7.9736,0>,T=0
HIT/VECTOR,<19.6545,8.3428,0>,<0.9205049,0.3907311,0>,<19.6545,8.3428,0>,T=0
HIT/VECTOR,<19.5676,8.712,0>,<0.9135455,0.4067366,0>,<19.5676,8.712,0>,T=0
HIT/VECTOR,<19.4741,9.0809,0>,<0.9063078,0.4226183,0>,<19.4741,9.0809,0>,T=0
HIT/VECTOR,<19.3737,9.4492,0>,<0.898794,0.4383711,0>,<19.3737,9.4492,0>,T=0
HIT/VECTOR,<19.2663,9.8167,0>,<0.8910065,0.4539905,0>,<19.2663,9.8167,0>,T=0
HIT/VECTOR,<19.1519,10.1832,0>,<0.8829476,0.4694716,0>,<19.1519,10.1832,0>,T=0
HIT/VECTOR,<19.0303,10.5487,0>,<0.8746197,0.4848096,0>,<19.0303,10.5487,0>,T=0
HIT/VECTOR,<18.9017,10.9129,0>,<0.8660254,0.5,0>,<18.9017,10.9129,0>,T=0
HIT/VECTOR,<18.766,11.2757,0>,<0.8571673,0.5150381,0>,<18.766,11.2757,0>,T=0
HIT/VECTOR,<18.6231,11.637,0>,<0.8480481,0.5299193,0>,<18.6231,11.637,0>,T=0
HIT/VECTOR,<18.4732,11.9966,0>,<0.8386706,0.544639,0>,<18.4732,11.9966,0>,T=0
HIT/VECTOR,<18.3162,12.3544,0>,<0.8290376,0.5591929,0>,<18.3162,12.3544,0>,T=0
HIT/VECTOR,<18.1521,12.7103,0>,<0.819152,0.5735764,0>,<18.1521,12.7103,0>,T=0
HIT/VECTOR,<17.981,13.064,0>,<0.809017,0.5877853,0>,<17.981,13.064,0>,T=0
HIT/VECTOR,<17.8029,13.4154,0>,<0.7986355,0.601815,0>,<17.8029,13.4154,0>,T=0
HIT/VECTOR,<17.6177,13.7645,0>,<0.7880108,0.6156615,0>,<17.6177,13.7645,0>,T=0
HIT/VECTOR,<17.4256,14.111,0>,<0.777146,0.6293204,0>,<17.4256,14.111,0>,T=0
HIT/VECTOR,<17.2265,14.4548,0>,<0.7660444,0.6427876,0>,<17.2265,14.4548,0>,T=0
HIT/VECTOR,<17.0206,14.7957,0>,<0.7547096,0.656059,0>,<17.0206,14.7957,0>,T=0
HIT/VECTOR,<16.8077,15.1337,0>,<0.7431448,0.6691306,0>,<16.8077,15.1337,0>,T=0
HIT/VECTOR,<16.588,15.4685,0>,<0.7313537,0.6819984,0>,<16.588,15.4685,0>,T=0
HIT/VECTOR,<16.3615,15.8001,0>,<0.7193398,0.6946584,0>,<16.3615,15.8001,0>,T=0
HIT/VECTOR,<16.1282,16.1282,0>,<0.7071068,0.7071068,0>,<16.1282,16.1282,0>,T=0
HIT/VECTOR,<15.8883,16.4528,0>,<0.6946584,0.7193398,0>,<15.8883,16.4528,0>,T=0
HIT/VECTOR,<15.6417,16.7737,0>,<0.6819984,0.7313537,0>,<15.6417,16.7737,0>,T=0
HIT/VECTOR,<15.3885,17.0907,0>,<0.6691306,0.7431448,0>,<15.3885,17.0907,0>,T=0
HIT/VECTOR,<15.1287,17.4036,0>,<0.656059,0.7547096,0>,<15.1287,17.4036,0>,T=0
HIT/VECTOR,<14.8623,17.7122,0>,<0.6427876,0.7660444,0>,<14.8623,17.7122,0>,T=0
HIT/VECTOR,<14.5893,18.0162,0>,<0.6293204,0.777146,0>,<14.5893,18.0162,0>,T=0
HIT/VECTOR,<14.3096,18.3155,0>,<0.6156615,0.7880108,0>,<14.3096,18.3155,0>,T=0
HIT/VECTOR,<14.0234,18.6097,0>,<0.601815,0.7986355,0>,<14.0234,18.6097,0>,T=0
HIT/VECTOR,<13.7306,18.8986,0>,<0.5877853,0.809017,0>,<13.7306,18.8986,0>,T=0
HIT/VECTOR,<13.4314,19.182,0>,<0.5735764,0.819152,0>,<13.4314,19.182,0>,T=0
HIT/VECTOR,<13.1258,19.4597,0>,<0.5591929,0.8290376,0>,<13.1258,19.4597,0>,T=0
HIT/VECTOR,<12.8139,19.7317,0>,<0.544639,0.8386706,0>,<12.8139,19.7317,0>,T=0
HIT/VECTOR,<12.4959,19.9976,0>,<0.5299193,0.8480481,0>,<12.4959,19.9976,0>,T=0
HIT/VECTOR,<12.1718,20.2573,0>,<0.5150381,0.8571673,0>,<12.1718,20.2573,0>,T=0
HIT/VECTOR,<11.8418,20.5106,0>,<0.5,0.8660254,0>,<11.8418,20.5106,0>,T=0
HIT/VECTOR,<11.506,20.7574,0>,<0.4848096,0.8746197,0>,<11.506,20.7574,0>,T=0
HIT/VECTOR,<11.1646,20.9975,0>,<0.4694716,0.8829476,0>,<11.1646,20.9975,0>,T=0
HIT/VECTOR,<10.8176,21.2308,0>,<0.4539905,0.8910065,0>,<10.8176,21.2308,0>,T=0
HIT/VECTOR,<10.4653,21.457,0>,<0.4383711,0.898794,0>,<10.4653,21.457,0>,T=0
HIT/VECTOR,<10.1077,21.676,0>,<0.4226183,0.9063078,0>,<10.1077,21.676,0>,T=0
HIT/VECTOR,<9.745,21.8876,0>,<0.4067366,0.9135455,0>,<9.745,21.8876,0>,T=0
HIT/VECTOR,<9.3774,22.0918,0>,<0.3907311,0.9205049,0>,<9.3774,22.0918,0>,T=0
HIT/VECTOR,<9.0051,22.2883,0>,<0.3746066,0.9271839,0>,<9.0051,22.2883,0>,T=0
HIT/VECTOR,<8.6281,22.477,0>,<0.358368,0.9335804,0>,<8.6281,22.477,0>,T=0
HIT/VECTOR,<8.2468,22.6579,0>,<0.3420201,0.9396926,0>,<8.2468,22.6579,0>,T=0
HIT/VECTOR,<7.8613,22.8308,0>,<0.3255682,0.9455186,0>,<7.8613,22.8308,0>,T=0
HIT/VECTOR,<7.4717,22.9955,0>,<0.309017,0.9510565,0>,<7.4717,22.9955,0>,T=0
HIT/VECTOR,<7.0783,23.152,0>,<0.2923717,0.9563048,0>,<7.0783,23.152,0>,T=0
HIT/VECTOR,<6.6812,23.3001,0>,<0.2756374,0.9612617,0>,<6.6812,23.3001,0>,T=0
HIT/VECTOR,<6.2807,23.4397,0>,<0.258819,0.9659258,0>,<6.2807,23.4397,0>,T=0
HIT/VECTOR,<5.8769,23.5708,0>,<0.2419219,0.9702957,0>,<5.8769,23.5708,0>,T=0
HIT/VECTOR,<5.47,23.6932,0>,<0.2249511,0.9743701,0>,<5.47,23.6932,0>,T=0
HIT/VECTOR,<5.0603,23.8069,0>,<0.2079117,0.9781476,0>,<5.0603,23.8069,0>,T=0
HIT/VECTOR,<4.648,23.9117,0>,<0.190809,0.9816272,0>,<4.648,23.9117,0>,T=0
HIT/VECTOR,<4.2332,24.0077,0>,<0.1736482,0.9848078,0>,<4.2332,24.0077,0>,T=0
HIT/VECTOR,<3.8162,24.0947,0>,<0.1564345,0.9876883,0>,<3.8162,24.0947,0>,T=0
HIT/VECTOR,<3.3973,24.1727,0>,<0.1391731,0.9902681,0>,<3.3973,24.1727,0>,T=0
HIT/VECTOR,<2.9765,24.2416,0>,<0.1218693,0.9925462,0>,<2.9765,24.2416,0>,T=0
HIT/VECTOR,<2.5542,24.3015,0>,<0.1045285,0.9945219,0>,<2.5542,24.3015,0>,T=0
HIT/VECTOR,<2.1305,24.3522,0>,<0.0871557,0.9961947,0>,<2.1305,24.3522,0>,T=0
HIT/VECTOR,<1.7058,24.3937,0>,<0.0697565,0.9975641,0>,<1.7058,24.3937,0>,T=0
HIT/VECTOR,<1.2801,24.426,0>,<0.052336,0.9986295,0>,<1.2801,24.426,0>,T=0
HIT/VECTOR,<0.8538,24.4492,0>,<0.0348995,0.9993908,0>,<0.8538,24.4492,0>,T=0
HIT/VECTOR,<0.427,24.463,0>,<0.0174524,0.9998477,0>,<0.427,24.463,0>,T=0
HIT/VECTOR,<0,24.4677,0>,<0,1,0>,<0,24.4677,0>,T=0
ENDSCAN
ENDMEAS/
| © 2024 Hexagon AB and/or its subsidiaries. | Privacy Policy | Cloud Services Agreement |